Research Focus
One central question that drives our research is to understand how a single species can overwhelm the host’s sophisticated immune system and cause serious disease in both plants and animals. The cross-kingdom nature of our research enables the integration of diverse research topics in addressing a few interesting questions.
2
How do two compartments of a genome interact with each to control the host-specific virulence of each strain? The discovery of accessory chromosomes (ACs) allowed us to divide a Fusarium oxysporum genome into the core and the accessory regions. To dissect the cross-talk between these two compartments of a genome, the Ma lab has studied key regulators – transcription factors (TFs)1 and kinases2–and found positive correlations between the proteome of an organism and the total number of TFs1 and protein kinases2 within it. This set up the foundation to search for master regulators used by both plant and human pathogens to coordinate the functions of ACs and the core genome as potential shared regulators controlling virulence for both plant and human pathogenic F. oxysporum strains, which can be used as novel targets.
3
What can Fusarium teach us about the mechanisms that underlie fungal adaptation to new hosts, from initial infestation to virulence factors?
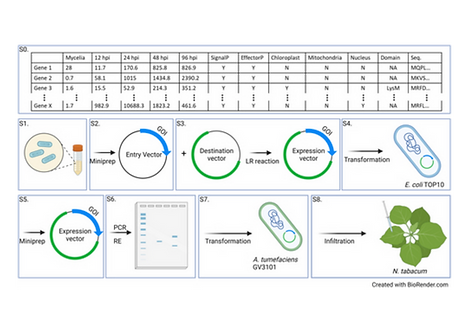
a) Effector Biology to test the hypothesis that Fusarium effectors facilitate the colonization and invasive growth in both plant and animal hosts.
b) Adaptation hotspots to test the hypothesis that there are genetic changes associated with distinct adaptation to the plant or animal hosts.
4
Translating basic research into novel approaches to control this cross-kingdom pathogen. To combat plant diseases, we will apply endophytes as potential bio-controls and introduce novel resistant genes into cultivated crops. Using our established high-throughput antifungal screens, we will develop lipid-based nanocarriers targeting the fungal cell wall to control infectious fusariosis. Ultimately, we aim to apply the gained knowledge to control this threatening cross-kingdom pathogen.
5
To study the evolution and diversity of Basil Downy Mildew pathogen Peronospora belbahrii, we will generate high-quality genome assemblies for new races of P. belbahrii combining PacBio single-molecule real-time (SMRT) long read sequencing technology with high coverage from Illumina sequence and investigate contributions of pathogen effectors using heterologous expression systems.